Faiss: Efficient Vector Similarity Search
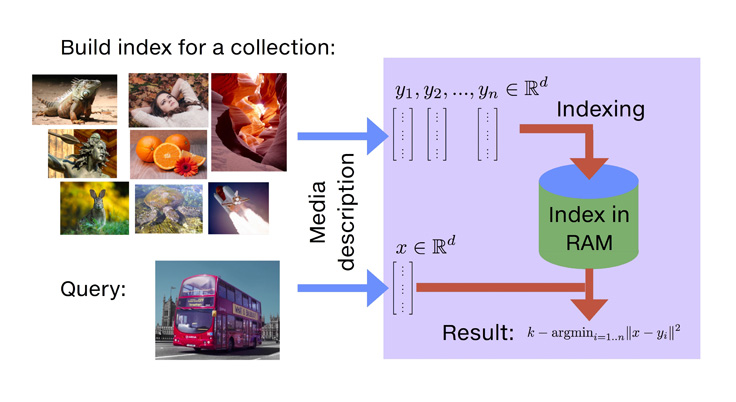
Introduction
As vector databases and similarity search become increasingly important in modern machine learning workflows, Faiss stands out as a robust solution for managing large-scale embeddings efficiently. These notes serves as a concise yet comprehensive reference for practitioners working on recommendation systems, image retrieval, NLP applications, and more. With Faiss, performing fast and scalable vector similarity searches is both practical and accessible.
What is Faiss?
Faiss (Facebook AI Similarity Search) is an open-source library developed by Meta for efficient similarity search and clustering of dense vectors. Optimized for high-dimensional data—such as embeddings from machine learning models—Faiss is capable of handling datasets with millions or even billions of vectors. Its primary applications include nearest neighbor search, recommendation systems, and duplicate detection.
- Why choose Faiss? Traditional brute-force search becomes impractical at scale; Faiss leverages advanced indexing techniques to deliver significant speed improvements.
- Advantages: High performance, memory efficiency, support for both CPU and GPU, and flexibility with multiple distance metrics (e.g., Euclidean, cosine).
- Considerations: Some index types provide approximate results and may require parameter tuning to achieve optimal accuracy.
Installation
Install Faiss and its dependencies.
CPU Version (recommended for most users):
pip3 install faiss-cpuGPU Version (for systems with NVIDIA GPUs and CUDA):
pip3 install faiss-gpuNote: Ensure your CUDA version is compatible with the Faiss release. Refer to the Faiss GitHub repository for details.
To verify your installation:
import faiss
print(faiss.__version__) # Example output: 1.8.0Install NumPy for vector operations:
pip3 install numpyCore Concepts
Before you start coding with Faiss, make sure you understand these key ideas:
- Vectors: These are lists of numbers (floats) that represent your data, like
[0.1, 0.2, 0.3]. Faiss works with NumPy arrays of typefloat32. - Index: This is the main structure in Faiss that stores your vectors and lets you search them quickly. There are different types of indexes, each with its own speed and memory usage.
- Distance Metrics: This is how Faiss measures how similar two vectors are:
- L2 (Euclidean): The default, good for most cases.
- Inner Product (IP): Used for cosine similarity (make sure to normalize your vectors first).
- Others: Like L1 or LInf, which you can set with
faiss.METRIC_*.
- Adding Vectors: Use
index.add(xb)to add your data to the index.xbshould be a NumPy array with shape(number of vectors, dimension). - Searching: Use
distances, indices = index.search(xq, k)to find the topkclosest vectors to your queriesxq. - Training: Some index types need to be trained first with
index.train(xb)before you can add data. This helps Faiss learn about your data.
Common variables you’ll see in examples:
d: The number of values in each vector (dimension), like 128.nb: How many vectors are in your database.nq: How many query vectors you have.k: How many nearest neighbors you want to find.
Common Index Types
Faiss has many indexes. Start simple, scale up. Here’s a table for quick comparison:
| Index Type | Description | When to Use | Pros | Cons |
|---|---|---|---|---|
| IndexFlatL2 | Brute-force exact search with L2 distance. | Small datasets (<10k vectors). | 100% accurate, no training. | Slow for large data. |
| IndexFlatIP | Exact search with inner product (cosine). | When cosine similarity is needed. | Simple, exact. | Same as above. |
| IndexIVFFlat | Inverted File: Clusters data, searches subsets. | Medium-large datasets. | Faster than flat, tunable accuracy. | Approximate; needs training. |
| IndexIVFPQ | IVF + Product Quantization: Compresses vectors. | Huge datasets, memory constraints. | Memory-efficient, fast. | More approximate, lossy compression. |
| IndexHNSW | Hierarchical Navigable Small World: Graph-based. | Real-time queries on large data. | Very fast searches. | Higher memory use, no built-in compression. |
| IndexLSH | Locality-Sensitive Hashing: Hash-based approx. | Binary vectors or quick rough searches. | Super fast for hashes. | Lower accuracy. |
For hybrids, combine like IndexIVFPQ for speed + compression.
Building and Training Indexes
Before building a Faiss index, it’s important to know whether your chosen index type requires training. The following flowchart summarizes the typical workflow for creating, training, and using a Faiss index:
flowchart LR
A(Create Index) --> B{{Training Needed?}}
B -- Yes --> C(Train Index)
B -- No --> D(Add Vectors)
C --> D
D --> E(Search)
Example: FlatL2 Index (No Training Required)
The IndexFlatL2 is the simplest Faiss index, performing exact nearest neighbor search using Euclidean (L2) distance. It’s ideal for small datasets or when you need 100% accuracy and don’t require any training phase.
import numpy as np
import faiss
d = 128 # Dimension
xb = np.random.rand(10000, d).astype('float32') # Database
xq = np.random.rand(10, d).astype('float32') # Queries
index = faiss.IndexFlatL2(d)
index.add(xb) # Add data
k = 5
distances, indices = index.search(xq, k) # Search
print(indices) # [[nearest IDs for query 1], ...]
print(distances) # [[distances for query 1], ...]Key Points:
- No training step is needed—just create the index and add your vectors.
- Best suited for datasets with fewer than ~10,000 vectors due to brute-force search.
- Returns exact nearest neighbors for each query.
Example: IVFFlat Index (Training Required)
The IndexIVFFlat index uses an inverted file system to cluster vectors, enabling much faster searches on medium to large datasets by searching only a subset of clusters. This index requires a training step to learn the cluster centroids before adding data.
import numpy as np
import faiss
d = 128 # Dimension
xb = np.random.rand(10000, d).astype('float32') # Database
xq = np.random.rand(10, d).astype('float32') # Queries
nlist = 100 # Number of clusters (tune for your data size)
quantizer = faiss.IndexFlatL2(d) # Base index for clustering
index = faiss.IndexIVFFlat(quantizer, d, nlist)
index.train(xb) # Train the index on your database vectors
index.add(xb) # Add vectors after training
k = 5
index.nprobe = 10 # Number of clusters to search (higher = more accurate, slower)
distances, indices = index.search(xq, k)
print(indices) # [[nearest IDs for query 1], ...]
print(distances) # [[distances for query 1], ...]Key Points:
- Training is mandatory before adding vectors—otherwise, you’ll get errors.
nlistcontrols the number of clusters; higher values can improve recall but may require more memory.nprobecontrols the number of clusters searched at query time; tune for your accuracy/speed tradeoff.- Suitable for datasets with tens of thousands to millions of vectors.
- Returns approximate nearest neighbors, but much faster than brute-force for large data.
Product Quantization (PQ) Compression in IVFPQ
Product Quantization (PQ) is a powerful technique used in Faiss to compress high-dimensional vectors, making it possible to store and search through massive datasets with limited memory. The IndexIVFPQ index combines inverted file indexing with PQ, enabling both fast and memory-efficient approximate nearest neighbor search.
PQ splits each vector into multiple sub-vectors (sub-quantizers), then quantizes each sub-vector separately using a limited number of bits. This reduces memory usage significantly, with a trade-off in search accuracy.
Example:
import numpy as np
import faiss
d = 128 # Dimension of vectors
nb = 10000 # Number of database vectors
nq = 10 # Number of query vectors
# Generate random database and query vectors
xb = np.random.rand(nb, d).astype('float32')
xq = np.random.rand(nq, d).astype('float32')
nlist = 100 # Number of clusters
m = 8 # Number of sub-quantizers (d must be divisible by m)
bits = 8 # Bits per sub-vector
quantizer = faiss.IndexFlatL2(d) # Base index for clustering
index = faiss.IndexIVFPQ(quantizer, d, nlist, m, bits)
index.train(xb) # Train the index
index.add(xb) # Add database vectors
k = 5
index.nprobe = 10 # Number of clusters to search
distances, indices = index.search(xq, k) # Search
print(indices) # [[nearest IDs for query 1], ...]
print(distances) # [[distances for query 1], ...]m: Controls how many sub-vectors each vector is split into. More sub-quantizers can improve accuracy but increase memory.bits: Number of bits used to encode each sub-vector. 8 is common for a good balance.
When to use: Choose IVFPQ when you need to scale to millions or billions of vectors and want to minimize memory usage, accepting some loss in search precision.
Normalization for Cosine
To perform cosine similarity search with Faiss, you must first normalize your vectors to have unit length. This ensures that the inner product between vectors is equivalent to their cosine similarity. Use the following approach to normalize both your database and query vectors:
import numpy as np
import faiss
# Generate random database and query vectors
d = 128
nb = 10000
nq = 10
xb = np.random.rand(nb, d).astype('float32')
xq = np.random.rand(nq, d).astype('float32')
# Normalize database and query vectors to unit length (L2 norm)
xb = xb / np.linalg.norm(xb, axis=1, keepdims=True)
xq = xq / np.linalg.norm(xq, axis=1, keepdims=True)
# Create an index for inner product (cosine similarity after normalization)
index = faiss.IndexFlatIP(d)
index.add(xb)
k = 5
distances, indices = index.search(xq, k)
print(indices) # [[nearest IDs for query 1], ...]
print(distances) # [[cosine similarities for query 1], ...]After normalization, use an index type that supports inner product search, such as IndexFlatIP, or set the metric to inner product for other index types. This workflow guarantees that the search results reflect cosine similarity rather than raw inner products.
Note: Always normalize your vectors before adding them to the index and before querying, otherwise the results will not represent true cosine similarity.
Saving and Loading Indexes
To ensure your Faiss index can be reused without retraining or rebuilding, you can persist it to disk and reload it later. This is especially useful for large datasets or production environments where index construction is time-consuming.
Saving an index:
faiss.write_index(index, 'my_index.faiss') # Save the index to a fileLoading an index:
index = faiss.read_index('my_index.faiss') # Load the index from a fileTip: Make sure to use the same Faiss version for saving and loading to avoid compatibility issues. If you are using GPU indexes, you may need to transfer the loaded index to the GPU after loading.
GPU Acceleration
Faiss provides seamless GPU acceleration when installed with the faiss-gpu package. To leverage GPU resources, transfer your CPU-based index to the GPU as follows:
import numpy as np
import faiss
# Create some sample data
d = 128 # Dimension
xb = np.random.rand(10000, d).astype('float32') # Database vectors
xq = np.random.rand(10, d).astype('float32') # Query vectors
# Build a CPU index
index = faiss.IndexFlatL2(d)
index.add(xb)
# Move the index to GPU
res = faiss.StandardGpuResources() # Initialize GPU resources
gpu_index = faiss.index_cpu_to_gpu(res, 0, index) # Move to GPU 0
# Perform a search on the GPU index
k = 5
distances, indices = gpu_index.search(xq, k)
print(indices) # [[nearest IDs for query 1], ...]
print(distances) # [[distances for query 1], ...]For multi-GPU setups or advanced GPU configurations, refer to the official Faiss documentation for detailed guidance.
Inspecting Indexes
Understanding the state and configuration of your Faiss index is essential for debugging, monitoring, and optimizing performance. Faiss provides several attributes and methods to help you inspect your index and verify its properties.
Key attributes to check:
index.ntotal: The total number of vectors currently stored in the index.index.d: The dimensionality of the vectors in the index.index.is_trained: Indicates whether the index has been trained (relevant for indexes that require training, such as IVF or PQ).- For IVF-based indexes:
index.nlist: The number of clusters (lists) used for partitioning the data.index.nprobe: The number of clusters to probe during a search, affecting the accuracy/speed trade-off.
Example:
print("Total vectors:", index.ntotal)
print("Vector dimension:", index.d)
print("Is trained:", index.is_trained)
if hasattr(index, "nlist"):
print("Number of clusters (nlist):", index.nlist)
print("Number of probes (nprobe):", index.nprobe)Reconstructing vectors:
To retrieve the original vector stored at a specific index (by its internal ID), use:
vector = index.reconstruct(0) # Retrieves the vector at ID 0
print(vector)Inspecting these properties helps ensure your index is correctly built, trained, and ready for efficient search operations.
Conclusion
Faiss is a robust library for fast vector similarity search, widely used in recommendation systems, information retrieval, and large-scale ML. By mastering its core concepts, index types, and best practices, you can efficiently manage datasets of any size. Whether you need exact or approximate search, CPU or GPU support, or memory-efficient compression, Faiss offers flexible solutions.